STATUS: RELEASED
Structural metadata are data about how a dataset or resource came about, but also how it is internally structured. How to FAIR
This step focuses on how to share your resource’s structural metadata - an explanation of what each piece of your data means and how it’s organised. Publishing it helps others understand, find and reuse your data more easily. This step also shows how to make the metadata readable by computers, which can support specific FAIR objectives you may already have.
Short description
Structural metadata (previously introduced in Metroline Step: Assess availability of your metadata), includes information about what data is collected and its format (e.g. “birth date” - YYYYMMDD), and, if applicable, the code systems and values applied. Similar to how an index at the beginning of a book refers to all its chapters and pages, it gives the reader an idea about the content and its definitions. For data resources, this is typically referred to as the codebook or data dictionary. You can find more information about how to generate one in Metroline Step: Analyse data semantics.
Once you’ve created this codebook, you should consider making it available to ensure clarity, usability, and trust in your dataset. This step will ensure that you can make a selection of data repositories that fit your research domain. Moreover, this step will point to tools that you can use to further increase FAIRness of your structural metadata.
Why is this step important
Registering (or publishing) structural metadata is crucial for ensuring the effective reuse and harmonisation of data structures across various research. This step enhances the contextual understanding of individual records, making data more meaningful and accessible, and improves the precision in data retrieval.
In other words, this step:
- Reduces ambiguity. Harmonises data elements, by providing definitions to each variable, making it easier to combine both existing and new datasets, as data alone might be meaningless or open to misinterpretation.
- Provides structural context. Explains how variables are related to one another and describes the format of the data.
- Increases findability. When variables are made publicly available, they allow more specific queries to be made about the resource.
- Enhances interoperability. Harmonised data definitions ensure that concepts are clearly and precisely described, enabling easier integration and combination across datasets.
- Enhances reusability. By clearly describing the data elements, the data can be reused in the future by yourself or by other projects with different research objectives.
- Improves reproducibility. Metadata about the structure of your data provides insight into how the original data was structured, even if the original data is no longer available.
Keep in mind your FAIRification objectives - registering structural metadata can facilitate the subsequent steps in the FAIRification process. For example, proper metadata is essential for applying Common Data Elements and creating a semantic model (Metroline Step: Apply common data elements, Metroline Step: Analyse data semantics).
How to
Step 1 - Prepare your codebook
For more information on how to choose which data elements to collect and how to create the codebook, see the following pages:Metroline Step: Apply common data elements, Create a Codebook and Metroline Step: Analyse data semantics.
The information you need to provide in your metadata are largely defined by the community you are part of and can vary significantly between research-domains.
- For instance, in microscopy research, an image is linked to details about the individual from whom a tissue sample was obtained. Here, characteristics such as age, sex, and diagnosis of that person serve as metadata about the sample, as well as the settings of the image itself, offering context about the microscopy image.
- However, in patient registry or clinical studies, this same information is considered the primary data, with their structural metadata including information about which variables are collected and the value ranges without specifying the individual record (for example, Age, captured as a integer ).
A group of Data Stewards conducted a campus-wide survey to assess current data management practices among their researchers. After collecting and preparing the data for sharing, they developed a codebook to document the dataset structure and definitions.
| Question | Variable_name | Variable_label | Value = 0 | Value = 1 | Value = 2 | Value = 3 | Value = 4 | Value = 5 |
|---|---|---|---|---|---|---|---|---|
| 1. Where are you employed? | Q001 | Where are you employed? | Radboud university medical centre (RUMC) | Radboud University (RU) | Donders Centre for Cognitive Neuroimaging | Donders Institute for Brain, Cognition and Behaviour | Other | |
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_1 | Audio or video recordings data | Not selected | Yes | ||||
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_2 | Behavioral and cognitive data | Not selected | Yes | ||||
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_3 | Genetic and/or genomic data | Not selected | Yes | ||||
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_4 | Health records data | Not selected | Yes | ||||
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_5 | Laboratory data | Not selected | Yes | ||||
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_6 | Medication data (i.e., pharmaceutical treatments) | Not selected | Yes | ||||
| 3. What type(s) of research data do you work with? (Multiple Choice) | Q003_7 | Medical Imaging data (e.g., tomography, infrared, magnetic resonance, ultrasound, x-ray) | Not selected | Yes |
| Q001 | Q003_1 | Q003_2 | Q003_3 | Q003_4 | Q003_5 | Q003_6 |
|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 1 | 1 | 0 | 0 |
| 1 | 0 | 0 | 1 | 1 | 1 | 0 |
| 1 | ||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 1 | 0 | 1 |
Step 2 - Select the most appropriate data repository for publishing
Once the codebook is ready, you can determine where and how it should be published. Structural metadata can be published in various forms, and the most suitable option often depends on the specific context of the research project and the FAIRification objectives. For certain projects, it might be practical to publish the codebook together with the research data in a repository, creating a cohesive and accessible package. However, in some projects where data cannot be published (e.g. due to privacy concern, legal restrictions, or intellectual property rights), it is appropriate to publish the codebook independently in a specialised catalogue, such as ART-DECOR. When selecting a repository or catalogue:
- Use domain-specific repositories where possible (a repository that is commonly used in your research domain). If you don’t know what repositories are used in your domain, you can make use of re3data or FAIRsharing to browse through repositories. Many filters can be applied (e.g., generation of a persistent identifier, selecting a country, data access level, etc.), making it possible to select a data repository that fits your needs.
- If there is no domain-specific repository, you can make use of generic or institutional repositories (for example, DANS Data Stations Life Sciences, Zenodo, Radboud Data Repository for Radboudumc researchers or DataverseNL for Amsterdam UMC, UMC Utrecht and Erasmus MC).
- Select a repository that makes it possible to publish documentation open access, even if the data is under restricted access. This way researchers can get an idea of what your data looks like, before actually needing to access the data.
- If possible, try to choose a repository that supports publishing metadata (or the codebook) in machine-readable formats, such as JSON/XML/RDF.
- Keep in mind the funder requirements – do they require you to publish in a specific repository?
- Some journals recommend specific repositories for researchers to use (e.g., PLOS One Recommended Repositories or Nature’s Scientific Data Repository Guidance).
The researchers decided to make use of the institutional data repository (Radboud Data Repository) to publish their data. Following best practices, they included the codebook alongside the dataset to enhance understandability.
Step 3 - Publish the codebook in the selected data repository
This step involves publishing the structural metadata in the selected data repository. Preferably the metadata is machine-actionable (see Step 4), but publishing your codebook as-is at this point may already suffice to reach your FAIRification objectives. Make sure that the codebook is published open access, even when the data is under restricted access. Some repositories that support this are DANS Data Stations Life Sciences (generic repository), Radboud Data Repository, and DataverseNL (institutional repositories).
The codebook was uploaded in a Data Sharing Collection from the Radboud Data Repository along with the dataset. This repository allows certain files in a collection to be flagged as documentation files. By marking the codebook as documentation, it is ensured that anyone downloading information about the dataset could immediately access and understand the content of the dataset, without needing to open the actual file.
Step 4 - Enhance findability and interoperability of structural metadata
This particular step might require onboarding of a data steward to assist with the process.
While stopping at Step 3 is already a good practice, ensuring that information about your data content is documented and made available, there are cases where you may want to further increase your FAIR maturity through additional mechanisms.
Findability
For surveys, patient registries, and other data types, it can be helpful to explicitly describe in the metadata what is actually being collected from the individuals (e.g., age, diagnosis, age at diagnosis). In such cases, platforms that present this information visually—like the one shown in the figure below—might offer a solution.
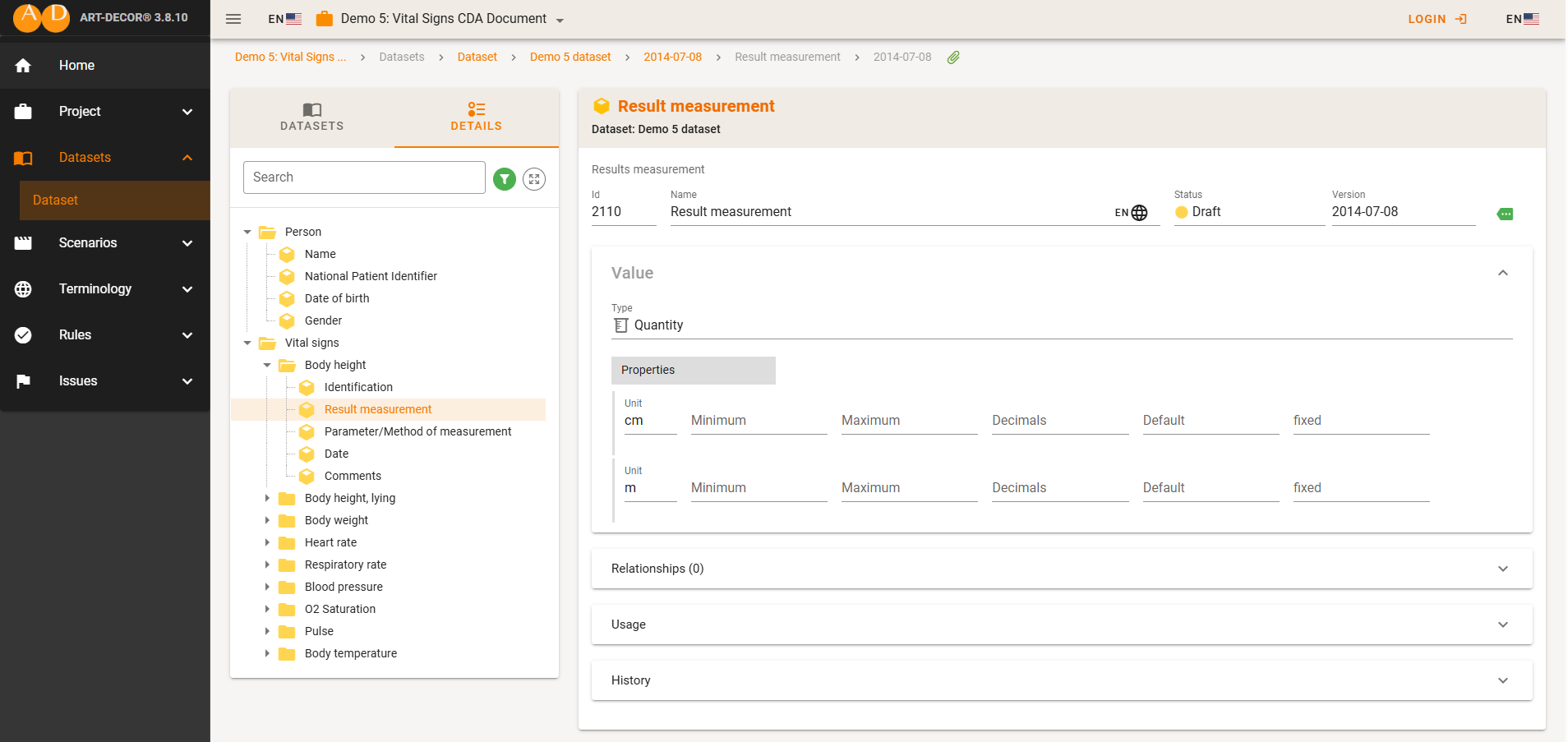
Depending on your field and data type, different tools may be available. Most of these tools support interoperability, which is also essential for FAIR data practices:
- Clinical studies and patient registries. ART-DECOR allows you to publish structured metadata about your dataset, including data elements in XML format (see example here).
- Patient Registries. ERDRI.mdr is a metadata repository that enables the publication of sampled data elements.
- Survey and/or any tabular datasets (rows and columns). DDI provides tools to convert your codebook into RDF or XML. Ideally, these files should be hosted in repositories that support these formats, but they can also be shared alongside your data following the guidance above.
While these are valuable solutions, it’s important to ensure that, if you share information about the content of your data in a different place than the location of your dataset, you reflect this on your metadata’s “single source of truth”, as explained in Metroline Step: Assess availability of your metadata. For example, if your institution connects with Health-RI to share metadata via a FAIR Data Point, make sure that the link to your structural metadata page is also included in that pathway.
Interoperability
Your original codebook file alone may not be sufficient to make your data interoperable. Two key characteristics are required:
- Machine-readable structure. Formats such as JSON (or JSON-LD), XML, OWL, or RDF help machines understand your data.
- Use of persistent identifiers. For example, using an ORCID to identify the dataset creator. This principle applies to other metadata fields as well:
| Concept | FAIR-term |
|---|---|
| Muscle Weakness | http://purl.obolibrary.org/obo/HP_0001324 |
To achieve interoperable metadata, consider using the following tools:
- FAIR Data Station. FAIR Data Station helps generate metadata templates for Biology and Omics Studies based on selected variables. It validates your codebook and produces an RDF file.
- CSV on the web. CSV on the web is a standard that describes the columns and rows of a given data file in csv. Therefore, it can be used across domains. The final format can be expressed in JSON or RDF and shared alongside your dataset.
If you have experience using other(s), please, leave a comment!
Spreadsheets, such as Excel files, are not inherently machine-readable and often require manual work for data cleaning and analysis. To improve interoperability, the Data Stewards generated machine-actionable metadata describing the dataset’s structure.
Using CSV on the web, a vocabulary designed to enhance metadata actionability, they created a JSON file that accurately reflects the content of the codebook. This effort significantly increased the dataset’s machine-readability and integration potential. Where possible, terms were mapped to ontologies. For example, Yes/No values were linked to:
- False: SNOMEDCT/64100000
- True: SNOMEDCT/31874001
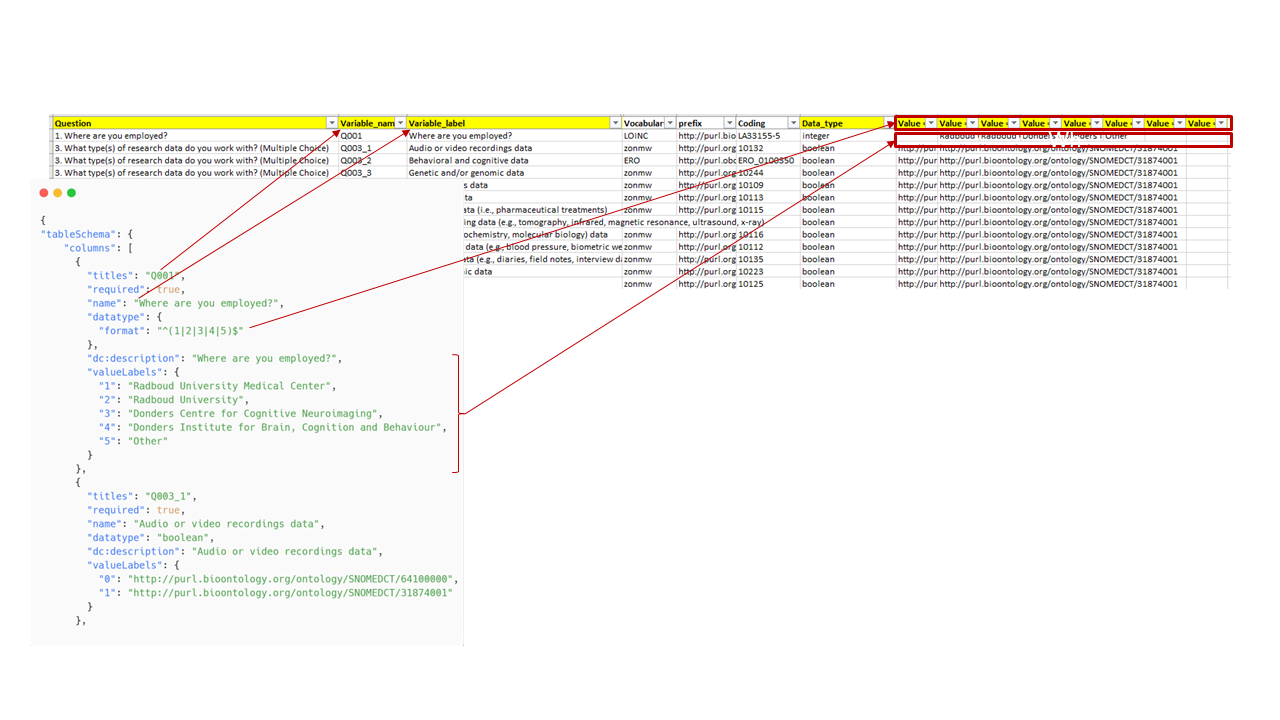
The next step is to make this JSON file available. It can remain within the same data collection or be hosted elsewhere, such as on GitHub or a JSON hosting service. However, if the file is moved to a location other than the the place where the dataset lives in, it is essential to ensure that this is documented in the organisation’s “single source of truth”, as already explained.
Guidance on maintaining metadata integrity can be found in the following Metroline resources: Metroline Step: Assess availability of your metadata and Metroline Step: Register resource level metadata.
Expertise requirements for this step
This step requires the following experts as described in the Metroline Step: Build the team step.
- Researcher. Provide the metadata/codebook.
- Data Steward. Find solutions and implement them or support the Researcher in the implementation.
Practical examples from the community
We’re looking for practical examples from the community to illustrate this step. If you have an example to share, we’d love to hear from you. Visit our How to contribute page to get in touch.
Training
Relevant training will be added soon.
Suggestions
This page is under construction. Learn more about the contributors here and explore the development process here. If you have any suggestions, visit our How to contribute page to get in touch.